How Design of Experiments enriches life sciences’ findings –

Design of Experiments (DOE) is a methodology misunderstood by many, understood by some, and actively used by even fewer than that. Wherever it does get used, though, it has the ability to completely transform scientific progress, as well as how scientists think about their work and understand what’s possible.
So, what is Design of Experiments (DOE)? At its most fundamental, it’s a framework for experimentation that allows us to do two things at the same time:
- Investigate the impact of multiple different factors, changed simultaneously, on an experimental process
- Investigate and explore the interactions between those factors
Seems straightforward, but these two abilities can let us derive incredible biological insight and identify the optimal conditions for an experiment or process in ways that more common experimentation methods can never hope to. DOE has the potential both to transform the power of our experiments and, more importantly, the depth of our insight.
The techniques for designing and analysing experiments that we use in DOE provide a practical framework for unpicking complex systems or processes—perfectly suited for the challenges we face in the life sciences. With DOE, scientists can take a more holistic approach in their methods when formulating and testing hypotheses.
Figure 1: how “OFAT” makes two passes to traverse the design space finding a local maxima, but missing the bigger picture… and the true maxima hiding elsewhere.
DOE stands in opposition to the old way of doing things: that is, by changing one factor at a time (OFAT). Because biological systems are characterised by complex interactions, changing one factor probably changes something else in the process. Biological variation, for example, can mean results vary randomly around a set point, even in a constant environment. Sample collection, transport, preservation, and measurement systems can introduce further sources of variation.
To take a more concrete example, it isn’t possible to fully understand the functional consequences of changing a protein’s structure without understanding all the contexts in which it appears. Its interactions within biological networks are what really define its function, so even minor changes can produce a plethora of unpredictable downstream and upstream effects. DOE allows the explorations of complex, multidimensional experimental design spaces, despite such methodological, biological, or chemical variations.
Where OFAT experiments ignore biology’s inherent complexity, it often identifies the wrong system state as the optimum (Figure 1). DOE instead handles this complexity directly.
Impact of DOE
Successfully landing DOE in a team, or an organisation, can have a major impact on the value of research. Here’s what that looks like:
Avoiding bias: DOE helps avoid unconscious cognitive bias and allows you to take a holistic approach when formulating and testing your hypotheses. You can be more confident in your data than you would have been with an OFAT approach; it’s all too easy to develop OFAT experiments that confirm hypotheses.
Confirming negative results: DOE means that you learn more about the space you are investigating than with OFAT, and you are also able to confirm if a negative result is a definitive negative result, giving you peace of mind when moving on to more promising avenues, instead of returning to the same old dead ends.
Factor screening: DOE allows you to identify factors that may have a material impact on a response and discard those that have little to no impact. Screening designs, such as fractional factorials, give you a lot of statistical bang for your experimental buck by carefully balancing the sets of conditions tested.
Lower cost, higher precision: DOE reduces the time, materials, and experiments needed to yield a given amount of information compared with OFAT. DOE also achieves higher precision and reduced variability, when estimating the effects of each factor and their interactions, than using OFAT.
The future of DOE: High Dimensional Experimentation (HDE)
A classic DOE design favours highly efficient designs, used over several iterations to explore a broad, albeit inexhaustive, design space. With HDE, we blow caution out of the water (Figure 2). When we have access to (and integration with) automation equipment that can run experiments with hundreds or thousands of runs, with volumes as low as 3-20 μl per run, we can very easily explore an entire design space in a single shot. (These are typically dispensers like the Dragonfly Discovery, the Gyger Certus Flex, and the Beckman Echo.)
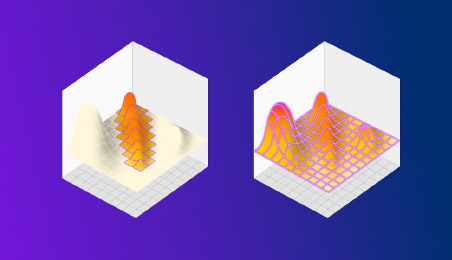
Figure 2: DOE on the left finds a better result than OFAT, but misses the true maxima hidden in an unexpected place. HDE instead explores the entire space in a single shot, revealing everything in one go.
Usually, the complexity of planning and running experiments like these would be completely prohibitive; the large volume of complex data and metadata alone can be daunting. However, with the right combination of hardware and software, it’s more than feasible. For truly successful HDE, though, it must include integrated experiment design and planning, as well as data capture and structuring.
Without the use of digital experiment software – platforms that handle experiment design, equipment automation, data aggregation, and structuring in a single place – HDE isn’t possible. But, in those R&D teams that are able to pull it off, the benefits are enormous:
- Dramatically faster assay development/method optimisation
- Full mapping of design spaces
- Lower operational cost and higher quality data
- Faster time-to-insight in optimised experiments
Those that remain wed to the familiar world of OFAT experimentation will probably continue to make incremental, linear improvements – even if they only identify false maxima. Those who have successfully adopted DOE, though, will make progress several orders of magnitude greater. But the few who can implement HDE? The sky’s the limit.
About the author

Markus Gershater, PhD, is co-founder of and chief scientific officer at Synthace. Originally, Gershater established Synthace as a synthetic biology company, but was struck by the conviction that so much potential progress is held back by tedious, one-dimensional, error-prone, manual work. Instead, he thought, what if we could lift the whole experimental cycle into the cloud and make software and machines carry more of the load? He’s been answering this question ever since. Markus holds a PhD in plant biochemistry from Durham University. He was previously a research associate in synthetic biology at University College London and a biotransformation scientist at Novacta Biosystems.
Source link
#Design #Experiments #enriches #life #sciences #findings