Cancer Research UK – Science blog
Cancer cells divide faster than healthy cells. Every time that happens, ecDNA can push them to evolve. ©Shutterstock/Christoph Burgstedt
We’re working out how cancers evolve. And we’re evolving our research to stop them.
We’ve known why cancers evolve for a long time: it’s the way they grow, survive and resist treatment. It’s a big part of why they’re so dangerous.
But the why isn’t enough. To stop cancers evolving, we need to understand how they do it.
Historically, that’s been a problem: the most aggressive cancers break some of the central rules of evolution.
When Charles Darwin described it almost 200 years ago, he said it was a gradual process. He’s been proved right. It took 6 million years for humans to evolve from our ape-like ancestors.
But some cancers can adapt to resist effective treatments in a week. The same slow story of step-by-step genetic mutations can’t explain both things.
For a long time, then, we’ve needed a new model. The fastest evolving cancers are often the ones with the lowest survival.
Now, thanks to Dr Paul Mischel, we have that model. His research has shown that cancers commonly use runaway circles of DNA called ‘extrachromosomal DNA’ (ecDNA) to evolve as quickly as possible. Since his discovery, our progress against it has started speeding up too.

Dr Paul Mischel’s team found the link between ecDNA and cancer evolution.
That’s also down to Mischel. He’s brought together a team of internationally recognised experts to tackle the problem, including Dr Howard Chang, a pioneer in understanding how DNA behaves in cancer.
Then, last year, the Cancer Grand Challenges initiative (which we co-founded with the National Institute of Cancer in the US) pressed the accelerator all the way down. Cancer Grand Challenges awarded Mischel’s team £20m in funding to take our knowledge even further. They’re not just chasing down this rule-breaking DNA: they’re adapting to it, evolving their methods so they can stop it in its tracks.
The disappearing cancer gene
‘Rule-breaking’ is a bit of an understatement. If these bits of cancer DNA were characters in a story, you’d call them devious. Chang compares them to the villains in James Bond movies. They want control, and they’ll do whatever they can to get it.
There’s more to that than helping tumours resist treatments, although that’s why researchers at Mischel’s clinic in California spotted them in 2014.
The team were trying to treat patients with aggressive brain tumours called glioblastomas. These tumours were all being driven to grow by the same gene (a short section of DNA), and the doctors had specific drugs to target it. They just didn’t work.
Looking for answers, the scientists zoomed in on the glioblastoma cells. Somehow, the cancer had removed almost all its copies of the vulnerable gene. Within a handful of generations, the tumour had adapted to grow without it.
It was as if children were being born that were completely unrecognisable from their parents. And if Darwin was right, that couldn’t make any sense.
So, the team kept investigating. They used these adapted cells to grow new tumours in the lab.
That revealed a whole new level of cunning.
Incredibly, the lab tumours looked like the original tumours did before treatment started. The vulnerable gene had quickly disappeared when threatened by cancer drugs and reappeared once it was safe for it to start causing damage again.
It’s not subject to the rules of normal cell division. That provides almost infinite adaptability.
Professor Charles Swanton, Cancer Research UK’s Chief Clinician
Mischel still calls the findings “frightening”. He’s a doctor and a scientist specialising in cancer. Just a couple of years before this, he had served as the President of the American Society for Clinical Investigation. He knew as much about what he was looking at as anyone else on the planet, and he had no way to explain it.
These are defining moments in science. The fast-evolving cells hadn’t just made the treatment fail; they’d shown that our understanding of cancer wasn’t working, either.
Vicious circles
So, what was going on?
Answering that requires a quick science lesson.
The cells Mischel’s team were looking at seemed to be ignoring the laws of inheritance, which come down to the way DNA is organised.
“When we think about inheritance, we think about chromosomes,” Mischel explains. These are tight coils of DNA containing lots of different genes. “We’ve got 23 pairs, and the reason we have them is to make sure that every cell in the body gets the same DNA.”
When a ‘mother’ cell divides, it completely replicates its DNA, so each ‘daughter’ cell receives the same 23 pairs of chromosomes. On rare occasions, this can go wrong in tiny ways, and over time there’s a chance these copying mistakes can build up into potentially cancer-causing mutations called oncogenes. These cause cells to divide more quickly, giving them chances to develop even more mutations.
But, even then, chromosomes in cancer cells don’t often crumple or fall apart. It should have taken many generations to cause the kind of changes Mischel’s team was seeing.
And yet the team’s cancer analysing tools told them chromosomes had to be responsible.
The most important of those tools were genomic cancer maps. They use our knowledge of human DNA to tie cancerous mutations to specific places on our chromosomes. The cunning one driving these glioblastomas was supposed to be on chromosome 7.
But there was another way to look. You can’t do this to help individual patients, but in some science labs it’s possible to locate bits of DNA in specific cells by watching them as they get ready to divide. When they did that, the team realised that their oncogenes weren’t where they were supposed to be.
In fact, they didn’t seem to be on the chromosomes at all. They were floating around them on rings of extrachromosomal DNA.
Off the map

Tiny circles of ecDNA are much harder to spot than chromosomes, but they can be key to understanding cancer.
These half-forgotten twirls of genetic information had been noticed – if not explained – before modern cancer maps came into use. But as the technology for tying genes to chromosomes advanced the field, ecDNAs fell into the background.
That was a mistake. In aggressive, treatment-resistant glioblastomas, ecDNA wasn’t a minor detail. It seemed to be central to what was going on.
Something about that made Mischel think about the solar system. For over 1,000 years, European and Arabic astronomers believed that the earth sat at the middle of it. That was thanks to the beauty and brilliance of a map by the Roman astronomer Ptolemy.
“All the measurements were right, but the map was wrong because they put the earth in the centre,” says Mischel. “And we realised, ‘My god, the cancer genome maps might be wrong’.”
Over the next few years, that’s exactly what the team showed.
“We proved that, in the most aggressive forms of cancer, the oncogenes aren’t where we thought they were, they’re actually on extrachromosomal DNA.”
Why is ecDNA such a problem?
Everything we’ve learned since Mischel’s discovery has shown that tackling ecDNA would make aggressive cancers much easier to treat. Ultimately, that would mean doctors could cure more cancers, and help people affected by them live longer, fuller lives. It’s why there’s a whole Cancer Grand Challenge dedicated to the task.
This is the equivalent of finding that master plan for cancer. All the important information is here.
Dr Howard Chang
This is what we know: ecDNA appears in 50% of cancer types, and it’s what gives many of them the ability to evolve so quickly. That’s because, when cells divide, ecDNA doesn’t neatly copy itself like well-ordered chromosomal DNA. One mother cell with 6 circles of ecDNA could split into one daughter cell with 12 and another with 0, one with 5 and one with 7, or any other combination.
“It’s not subject to the rules of normal cell division,” explains Professor Charles Swanton, our chief clinician. “It’s essentially random and unpredictable.
“So, what ends up happening is that these ecDNAs can wax and wane. They can almost completely disappear from the tumour and then come back after you release it from drug pressure. That provides almost infinite adaptability, which means that drugs that target [genes on ecDNA] are likely to fail.”
The big reveal
All that makes ecDNA an extremely complicated problem, but eDyNAmiC, the Cancer Grand Challenges team led by Mischel, is making progress on it at a dizzying pace.
In less than a year, they’ve come up with a way of improving our cancer maps.
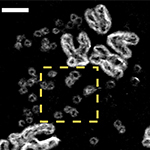
eDyNAmiC have developed a way to locate cancer genes on ecDNA.
The old ones looked at all the DNA in a patient’s tumour at once, so they could only point to where cancerous mutations were supposed to be on chromosomes. No one realised until Mischel’s discovery, but this meant ecDNAs could sneak around the edges like spies.
It was as if the ecDNAs were using code machines to scramble the messages scientists were reading from cancer cells. It was possible to see what they said, but not where they came from.
With its new system for separating DNA by size as well as content, eDyNAmiC has cracked that code. Because ecDNA is much smaller than chromosomal DNA, this approach enables researchers to use all their high-tech equipment to analyse ecDNA by itself.
As a result, we can finally see the truth about ecDNA.
In essence, in many of the most aggressive and treatment resistant cases, the ecDNA is the cancer.
“The chromosomal version of the gene is actually normal,” explains Chang. “You can say, ‘If I just get rid of ecDNA, this cell wouldn’t even know it’s necessarily cancerous.’”
So, in the most treatment-resistant cancers, it looks like ecDNA could be more than just another villain. We may have found the criminal mastermind.
“If you watch a Bond movie, or something like it, there’s a point where the villain starts explaining the grand plan,” Chang continues. “They say, ‘This is how I’m going to take over the world!’ And then you’re like, ‘Okay, that’s why all these things are happening!’.
“This is the equivalent of finding that master plan for cancer. All the important information is here.”
Adapting to ecDNA
As our understanding of ecDNA becomes clearer, we’re getting closer to working out how we can stop it. This latest finding suggests that we could treat a range of different cancer types with one treatment that targets the unique features of ecDNAs. Already, eDyNAmiC have identified a possible drug candidate.
Both Mischel and Chang are clear on what’s making this possible. “We simply could not bring this extraordinary group of people together to do this if it were not for the Cancer Grand Challenges programme,” stresses Mischel. “It’s a very fresh, interesting, forward-thinking and powerful way to do the kind of science that needs to be done to solve and tackle big problems.”
Their work is even linking into our flagship lung cancer study, TRACERx, which is run by Swanton. Not only is that the biggest project of its kind in the world, it’s also specifically about ‘TRAcking Cancer Evolution through treatment (Rx – the medical abbreviation for ‘prescription’).
The next phase of TRACERx is called TRACERx EVO. That could explain the Cancer Grand Challenges, too. We’re not just working on evolution: we’re evolving ourselves.
Tim
Source link
#Cancer #Research #Science #blog